| FAQs |
|---|
|
1. What is the relationship between PDBbind and PDBbind-CN database? The PDBbind database was orginally developed by Prof. Shaomeng Wang's group (http://sw16.im.med.umich.edu) at the University of Michigan in USA, which was first released to the public in May, 2004. This database is now maintained and further developed at this web site, i.e. PDBbind-CN, by Prof. Renxiao Wang's group (http://www.sioc-ccbg.ac.cn) at the Shanghai Institute of Organic Chemistry, Chinese Academy of Sciences in China under a mutual agreement with the University of Michigan. All registered users of the original PDBbind database have been granted access to the PDBbind-CN web site as well. |
|
2. Do I need to register before using the PDBbind-CN database? The basic information of each complex in PDBbind is totally open for browsing. Users are however required to register for access under a license agreement in order to utilize the full functions provided on this web site or to download the contents of PDBbind. The registration is free of charge to all academic and industrial users. Please go to the "[REGISTER]" page to register. Technical support contact with: liuhai@mail.sioc.ac.cn |
|
3. Do I need to pay for using the PDBbind-CN database? No, the PDBbind-CN database is a totally free and open access database, we only require users to register before use. |
|
4. Can I redistribute data in the PDBbind-CN database? No, please refer to our user license agreements, any kind of redistribution of the PDBbind-CN database is strongly prohibited. You can only make links to our database unless you get our authorization. |
|
5. What is the recommended software environment for browsing the PDBbind-CN website? Viewing of 3D molecular structures is powered by ChemAxon. This requires you have Java JRE v1.50 or above installed on your computer. Substructure search is powered by Open Babel. Microsoft's Internet Explorer (6.0 or higher) is recommended for browsing this web site. |
|
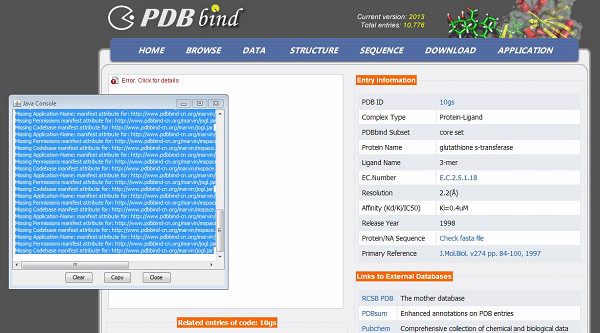
6. I can not load the Java Applet plugin in Windows 7 64-bit operating system
If you see the error messages like below: |
|
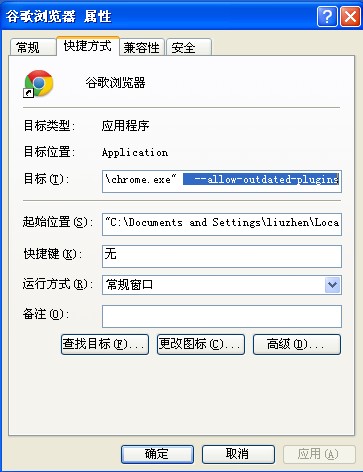
7. How to close the endless Java Applet plugin outdated prompts in Chrome or Firefox browsers?
Yes, sometimes the Chrome or Firefox browsers may prompt the Java Applet plugin is outdated, or ask you to run it once or run it always, or need to install plugin in Linux system. You can try to add a parameter to the browser executive file: |
|
8. How to cite the PDBbind-CN database?
[1] Minyi Su, Qifan Yang, Yu Du, Guoqin Feng, Zhihai Liu, Yan Li,* Renxiao Wang,*, "Comparative Assessment of Scoring Functions: The CASF-2016 Update", J. Chem. Inf. Model, 2019, Vol. 59: pp 895-913.(CASF-2016) |








 times since Nov 2007.
times since Nov 2007. 
Copyright ©2007-2025 上海盈赛思信息科技有限公司 网站备案号:沪ICP备2021015625号-3 ![]() 沪公网安备:正在申请中
沪公网安备:正在申请中
Technical Support(技术支持): yingsaisi@foxmail.com